
McClain is known for the dissection of RNA-protein recognition primarily through genetic studies in bacteria. The work is meticulous, self-critical and has led to new ideas.
Work on multimeric precursor tRNA processing to tRNA in the 1970s remains a paradigm of RNA synthesis. He defined a seven-step pathway leading from transcribed DNA to large RNA intermediates that accumulated in successions of mutant cells lacking germane processing enzymes and whose nucleotide sequences defined the ordered steps, including formation of tRNAs' 3'-CCAOH end for amino acid acceptance. Other precursor tRNA processing work continued.
The alanine tRNA and aminoacyl-tRNA synthetase system is an example of his computer-guided mapping of tRNA sequence onto amino acid acceptor function. First, a G-U wobble base pair in the acceptor helix of alanine tRNA emerged as a key computer correlate of alanine acceptor specificity. Then, in mid 1988, the substitution of this G-U pair in a gene for a different tRNA changed the acceptor specificity of the transcribed tRNA substantially to that of alanine. Armed with new genetic and biochemical data, McClain hypothesized later in 1988 that the G-U pair determines alanine tRNA acceptor specificity by inducing a local helical irregularity, a novel concept. The tRNAs for all other amino acids lack this irregularity, providing the structural basis for their exclusion by the alanine synthetase enzyme. Subsequent work (Choi et al., 2002) revealed that the G-U pair is but one member of a nested group of base pairs in the alanine tRNA helix that synergistically cooperate to form the key recognition structure for the synthetase enzyme. Cooperation between multiple nucleotides in recognition was unexpected and novel, and cannot be predicted by single-mutation or 3-D structural analyses. Eventually, Dr. Shigeyuki Yokoyama determined the crystal structure of a complex between alanine tRNA and its aminoacylating enzyme (Naganuma et al., 2014). This structure confirmed all the work from 1988 through 2002, in particular the acceptor helix irregularity, which takes the form of a substantial widening of all the base pairs in this helix, predictably widened via synergistic nucleotide cooperation, to avoid a steric clash of the backbone of prebound tRNA with the synthetase enzyme.
© 2016
Forward
Transfer RNA (tRNA) converts the instructions in a gene into amino acid subunits, forming protein. Proteins are composed of 20 amino acid types, wherein each amino acid type corresponds to one type of tRNA. The tRNA is built of nucleotide subunits. The sequence of nucleotides in a tRNA is highly variable and determines the specificity of each tRNA for its amino acid.
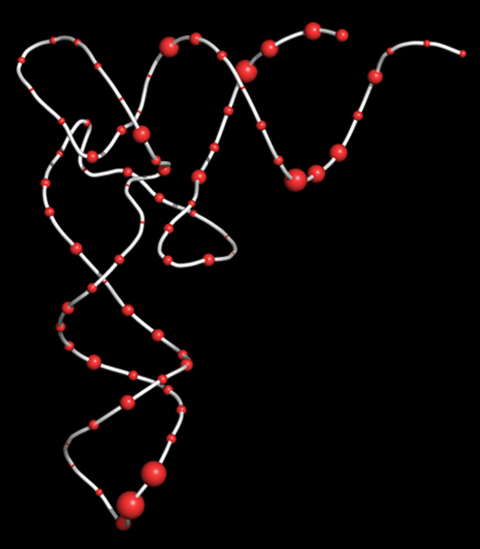
Representation of the common 3-D structure of tRNA, comprised of a repeating backbone (white) with one of four pendent nucleotides at 76 sites (red spheres). Our original computer analysis of the nucleotide sequences of the 20 types of tRNA enabled us to predict which nucleotides were both favored by each tRNA acceptor type and disfavored by the other 19 tRNAs. Larger red spheres were more predictive. Guided by computer analysis, we chemically redesigned a tRNA gene from type 1 to type 2. Protein sequencing revealed that cells containing the redesigned tRNA switched their specificity to that of type 2.
Formal Education
- B.S., 1964, Iowa Wesleyan College.
- Ph.D., 1968, Purdue University.
Mentor, Sewell P. Champe. - Postdoctoral Research, 1967,
Purdue University. - Postdoctoral Research, 1969,
Medical Research Council,
Laboratory of Molecular Biology Cambridge, England.
Mentors:
Sydney Brenner, Francis Crick
and Fred Sanger.
- Faculty Appointment, 1971,
University of Wisconsin-Madison. - Contact Information:
- University of Wisconsin-Madison
Microbial Sciences Building
1550 Linden Drive
Madison, WI 53706, U.S.A.